3. Command-line in Unix-Like Consoles Basics (In Browser)
Command-line/BASH
Learning Objectives
This module is meant to be a broad and very basic introduction to the command-line. Do you need this module? Well, it depends, but one way you might know is that you are told you need to sign-in to the server, such as a high-performance computer (HPC). So you’ll usually know that you need it. If you don’t know that you need command-line, You will learn enough to be functional, but many aspects will be missed. We have modules where provide more detail, but they require a Unix-based computer (like Mac), and this module leverages a web-interface: Jupyter Labs. This is done because many users will be on Windows-based machines and may not have access to a Unix-Linux based computer. However, there are some simple things you can learn and gain a lot of functionality.
To start a Jupyter Terminal, we will start with Jupyter Labs. We can then open a new terminal.
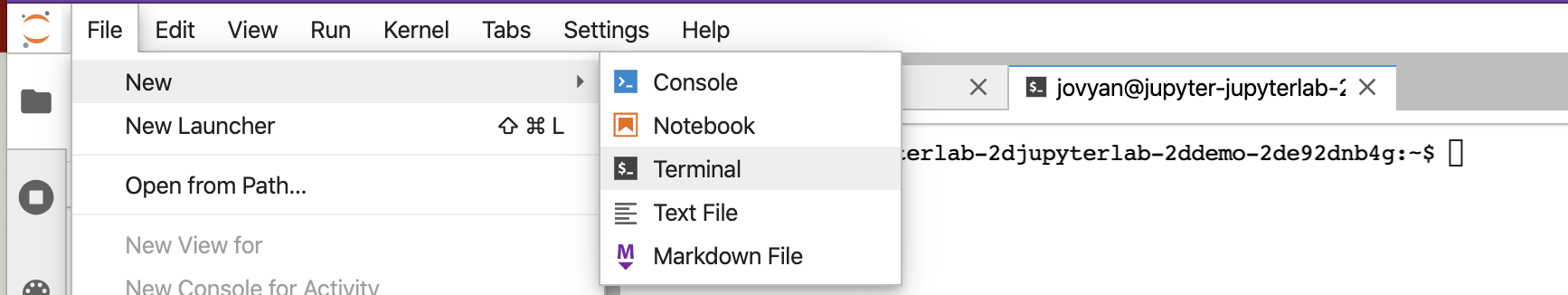

We see something similar to below.
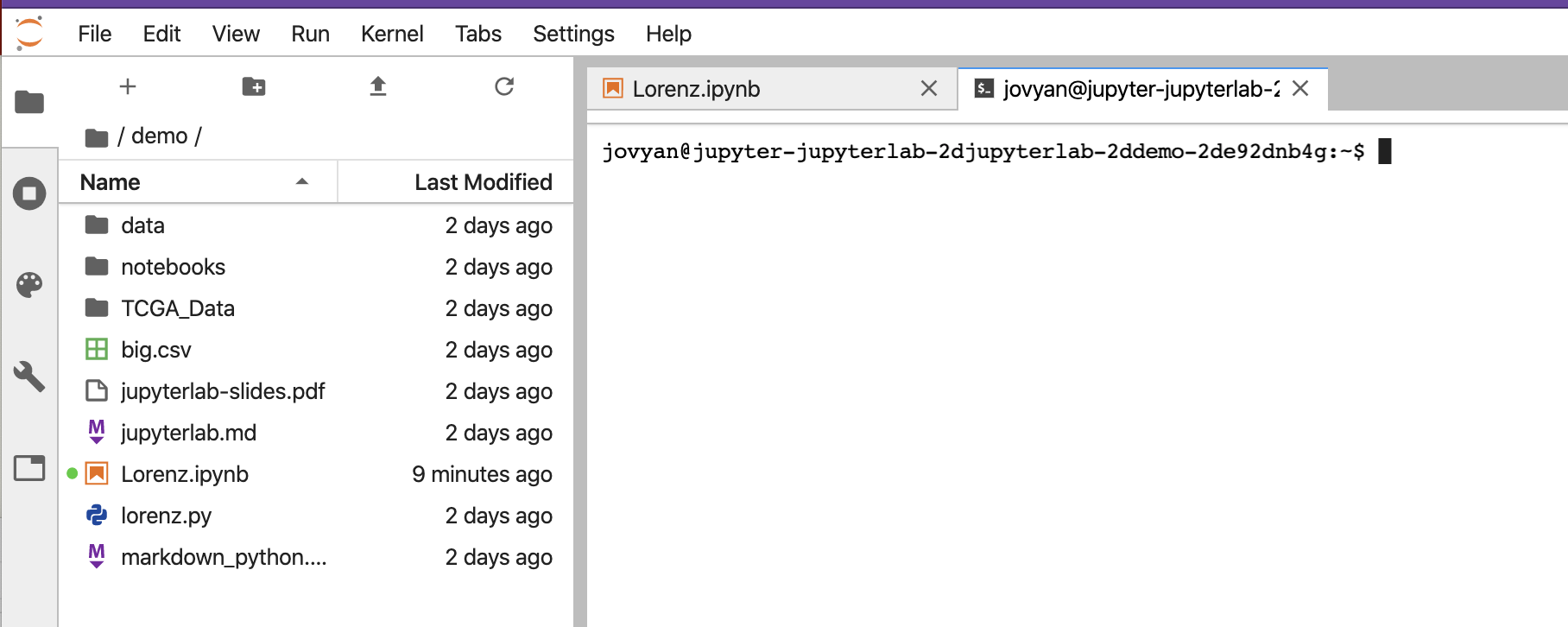

On the left above is a file structure with several starter files in it. On the right is the terminal that we will learn using.
High-level: What is Unix/Linux/Bash/Command-line?
These are terms that are used loosely, and not really reflective of their technical meaning. If I said to you this course teaches Windows for example, for most something comes to mind. However technically, Windows could be Windows 95,98,3.1, ME, XP, and so forth. Same here – just more extreme. Learning command-line is generally the same as learning BASH (the most common shell environment), and the same as learning to use Linux/Unix computers for most beginners. Down the road, there are important distinctions, so to keep all of this accessible to new users, we will not go down these rabbit holes.
Part 1: Navigating and commands
Unix, Linux, and Command-line For Bioinformaticians
Overall, this module was developed originally for the command-line and is being ported over to Jupiter. We do still have command-line instructions, and at some level, its good to see those since so many recipes presume the command-line. That said, it should always be straight forward to adapt these to Jupytor.
UNIX is an operating system around in the 60s and broadly refers to a set of programs that all work the same way at the command-line. They have the same feel. They have the same philosophy of design. Ok, it’s a specific operating system owned by AT&T, however, these days it refers to a program that all follow a common framework. There are many types of Unix – MacOSX, Linux, and Solaris where each of those is essentially different sets of codes owned by different companies or groups to get the common Unix common framework. MacOSX is owned and developed by Apple. Solaris is owned by Sun and Oracle. Linux is open-source and built from a community-led by Linus Torvalds, and was meant to work on x86 PCs. The x86 refers to a type of CPU architecture used across most personal computers today (both Mac and PC). If I log into a Unix machine in 1980, 1990, 2000, 2010, 2017 – it will often feel and work the same. By comparison, Windows is not Unix. Even if you go to the command-line everything is different and changes over the years.

Windows over 20 years
Windows has generally seen many different Graphical User Interfaces (or GUI’s) over the years. Generally, the advent of Windows and key innovation was to remove the need for shell level computing. However a lot of scripting and ability to manipulate, wrangle, edit, and work with text was lost. Bioinformatics tends to need these tools, and these are generally found in the various flavors ‘nix. There are ways to get to a ‘nix environment within Windows such as through Cygwin or other VM devices but we generally don’t discuss that in this material.

Command-line shells
Command-line shells are started up from a terminal program. Every Mac computer has Terminal preloaded. Start that up and you’ll see a prompt from the shell. The shell is actually a program that responds to you, and you can change its look and feel. Most people like the shell that is called bash. With Catalina, there is a recommendation of using zsh. However, 15 years ago, tcsh was more common. There are others like c-shell (csh) and ksh.
Bash is pretty handy in that things like up-arrow takes you the previous command and you can press ‘tab’ to autocomplete. Now an important thing is that when bash starts .bash_profile is executed for login shells, while .bashrc is executed for interactive non-login shells. We can store a lot of settings here. Settings for the shell are also called environmental variables. You can see some examples such as typing echo $HOME' where echo simply prints the variable. $PATH is really important because any program normally has had the full name or path to run it. However, those in the path don’t. For example, let’s say the program ‘ls’ is stored in /usr/bin/ls. To run it, you’d have to type /usr/bin/ls. However if add /usr/local/bin to our path, then we only have to type ls. We can have a lot of things in our path, and then separate them by colons. If you like, type echo $PATH to see what’s in your current path. All of your favorite startup settings are in your .bashrc file. In some cases, a default setup is only calling up .bash_profile. Here, people usually have only one command in it – that is to source .bashrc.
Directories are something we’ve touched on, but its important to know that every file is within a directory. In Unix, these are separated by “/”.. If we cd to the top level it would be cd /. The character ~ has a special meaning and it means the home directory. Typing cd ~ changes directory to our user’s home. you can do that followed by pwd to figure out your home directory.
There are some conventions. The bin the directory is typically where you put executable programs. So the first good thing to do is to create a bin in your home directory. These are often called local executable files. To actually make the bin meaningful, you’d have to add it to your path, such as set PATH=$PATH:~/bin, would add that local bin to your existing path. There are going to some programs installed as superuser or root (who can read and write anywhere. These are typically in /usr/bin. The directory /etc is where settings are – and don’t go here unless you know what you are doing. Don’t worry – you shouldn’t be able to do anything without becoming superuser.
There are many different resources for learning command line on Linux/Unix based systems. Typically, a user may need to know 20 to 40 commands, with cd, ls, less being common. We have provided a 1 cheat sheet below and we link to some provided by others. All of the commands have lots of options, and one can learn about them by typing ‘man command’, ( ‘man grep’ for example). However, most people just google Linux command options. It is important to know that there are thousands of Linux commands, but most people only remember a small subset that is specific to their field. Some example resources are:
- https://learncodethehardway.org/unix/bash_cheat_sheet.pdf
- https://files.fosswire.com/2007/08/fwunixref.pdf
- https://www.cheatography.com/davechild/cheat-sheets/linux-command-line/
- https://www.tjhsst.edu/~dhyatt/superap/unixcmd.html
Cheatsheets
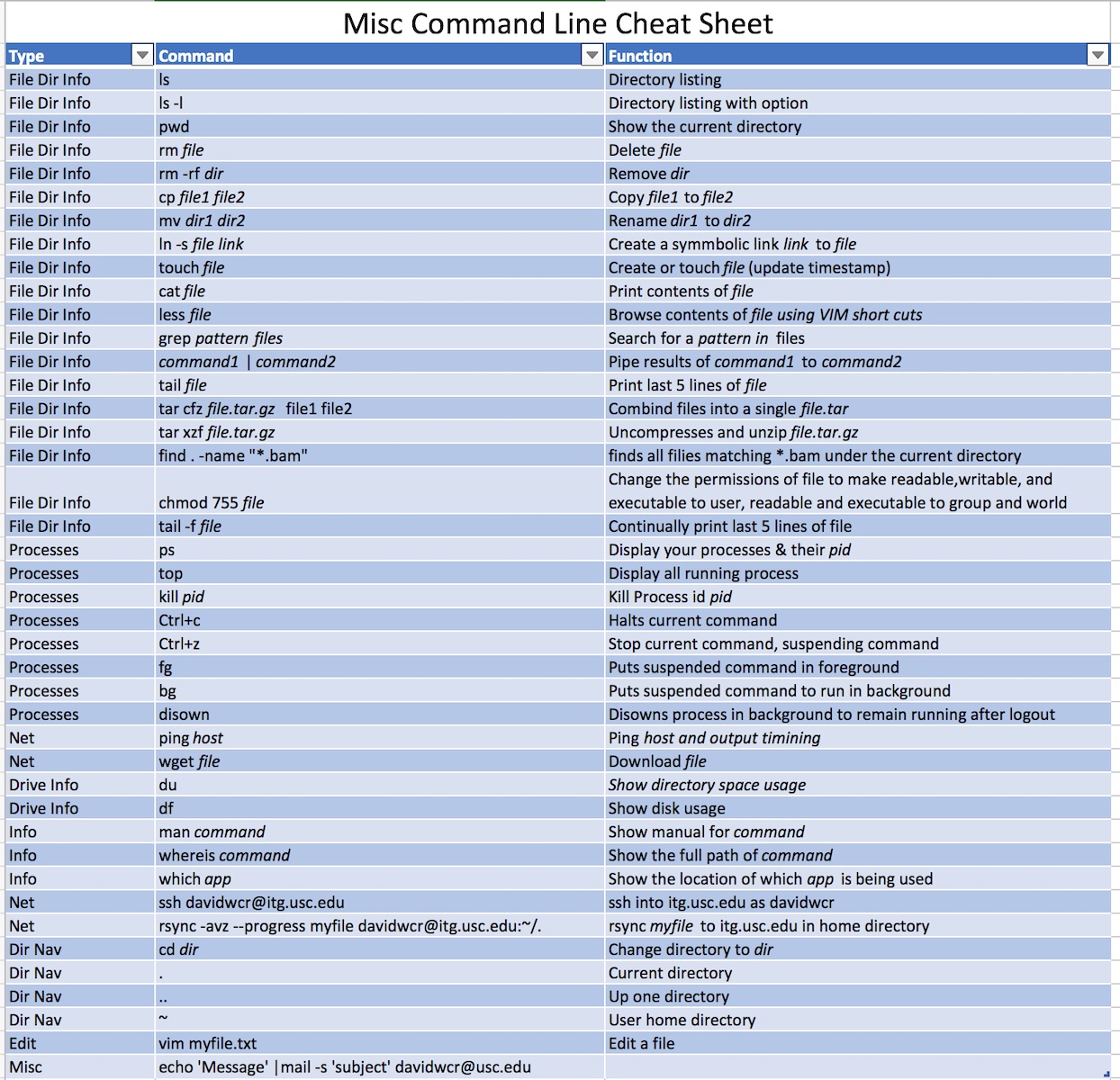

Part 1: Navigating and managing files
A few important examples. First to go up a directory, and then list the contents including permissions.
!cd .. !ls -l
Change to a directory in the users home directory
!mkdir mydir !cd ~/mydir
Basics: moving around in directories
These are the basic commands needed to move around in directories.
pwd # Get full path of the present working directory (same as "echo $HOME") ls # Content of pwd ls -l # Similar as ls, but provides additional info on files and directories ls -a # Includes hidden files (.name) as well ls -R # Lists subdirectories recursively ls -t # Lists files in chronological ordercd <dir_name> cd # Brings you to the highest level of your home directory. cd .. # Moves one directory up cd ../../ # Moves two directories up (and so on) cd - # Go back to you were previously (before the last directory change)
Basics: Files and directories
mkdir <dir_name> # Creates specified directory rmdir <dir_name> # Removes empty directory rm <file_name> # Removes file name rm -r <dir_name> # Removes directory including its content, but asks for confirmation,'f' argument turns confirmation off cp <name> <path> # Copy file/directory as specified in path (-r to include content in directories) mv <name1> <name2> # Renames directories or files mv <name> <path> # Moves file/directory as specified in path
Pipe | and Redirect >
One of the most important parts of Unix is pipe and redirect. Piping is done using the | symbol, and sends the output from what’s left of the pipe, to what’s right of the pipe. Redirect > puts the output of the left into the file in the right.
An example of a pipe that takes our history and pipes it to grep which only prints lines that match.
history | grep ls
An example of redirecting that output to a file
history > myhistory.txt
Permissions
Permissions are an important early concept. A simple explanation is that files can be readable(+4), writable(+2), and executable (+1) to yourself, your group, the world (that can login), and in that order. If something is read-only, it’s 4. If something is read and writable, its 6. If something is readable and executable its 5. A script needs to be executed, and thus it’s possible to make a script executable using the chmod command, with 7 for the user, 5 for the group, and 5 for the world.
To show this example, lets first get a script called myscript.sh using the linux command `wget` that is provided at the url https://itg.usc.edu/site/myscript.sh. As a note the command wget is handy for scraping web-pages. The script as we learn is simple, and starts with a shebang or declaration of what shell should run the following lines.
#!/bin/bash
# declare STRING variable
STRING=”Hello World”
#print variable on a screen
echo $STRING
We provide that just FYI, because we will just download it from a site
wget --no-check-certificate https://itg.usc.edu/site/myscript.sh
You’ll see something like:
--2020-04-23 12:59:11-- https://itg.usc.edu/site/myscript.sh Resolving itg.usc.edu (itg.usc.edu)... 128.125.215.226 Connecting to itg.usc.edu (itg.usc.edu)|128.125.215.226|:443... connected.
This will have put the script in your directory. But you cannot run it because its not executable. to see this type:
ls -l
We see
jovyan@jupyter-jupyterlab-2djupyterlab-2ddemo-2dz7wzm2in:~$ ls -l total 44 -rw-r--r-- 1 jovyan jovyan 1323 Apr 22 04:17 appveyor.yml drwxr-xr-x 1 jovyan jovyan 4096 Apr 22 04:17 binder drwxr-xr-x 1 jovyan jovyan 4096 Apr 22 04:17 data drwxr-xr-x 1 jovyan jovyan 4096 Apr 23 13:00 demo -rw-r--r-- 1 jovyan jovyan 2653 Apr 22 04:17 LICENSE -rw-r--r-- 1 jovyan root 100 Apr 23 12:54 myscript.sh -rw-r--r-- 1 jovyan jovyan 3106 Apr 22 04:17 README.md -rw-r--r-- 1 jovyan jovyan 2162 Apr 22 04:17 talks.yml -rw-r--r-- 1 jovyan jovyan 6275 Apr 22 04:17 tasks.py
This gives the permissions as we will show. We now must make it executable
chmod 755 myscript.sh ./myscript.sh
This prints
Hello World
Concept: File permissions & chmod
Lets dive in.
You are user. A linux computer expects multiple users and they form the world of users. You can be assigned to a group or groups. Other users on the computer may be in your group, and there are some groups you don’t belong to.
Level 1: You the user, and your permissions. Perhaps your username is john_doe. If you want to be able to view a file, give yourself a point (+4). If you want to be able to write to a file or change a file, give yourself four points (+2). If you want to be run a file – such as a script as a program, give yourself a point. Want to do all 3? 3+2+1=7.
Level 2: Groups. Perhaps you belong to bioinformaticians. If Jane_doe is also a bioinformatician. Do you want Jane to execute a file? Well, she technically must be able to read it. So she needs 4 + 1 points = 5 points
Level 3: World. What about others on the computer? What if you don’t want others to be able to read or execute – they get 0 points.
Let’s look at an example. In your Jupyter notebook, type
ls -l
This asks to list the files in our current directory, showing all permissions, dates and so forth. My example is below:
total 1112 -rw-r--r-- 1 jovyan jovyan 8 Apr 15 06:28 apt.txt -rw-r--r-- 1 jovyan jovyan 226 Apr 15 06:28 environment.yml -rw-r--r-- 1 jovyan root 1116435 Apr 17 03:37 index.html -rw-r--r-- 1 jovyan jovyan 4326 Apr 17 03:38 Index.ipynb -rwxr-xr-x 1 jovyan root 19 Apr 17 03:35 ls.sh
Above, we see permissions. We see the apt.txt can be read and edited (write) by the level 1 user.
-rw-r--r-- 1 jovyan jovyan 8 Apr 15 06:28 apt.txt
It can be read by users who are level 2 and 3. If you shared the computer with other users, this may be them.
The last file can be run and is executable because of the x. In fact, we see that everyone on the computer can run it:
-rwxr-xr-x 1 jovyan root 19 Apr 17 03:35 ls.sh
What if I just want only myself to be able to run the file? I would chmod 700 ls.sh and then it becomes
-rwxr-xr-x 1 jovyan root 19 Apr 17 03:35 ls.sh
So to set permissions for User, Group, World – that would be 750
chmod is the command, chmod 700 myfile.txt would do it!
Make files executable via chmod 700
What command will we use time and again? We will make scripts, and we need them to be able them to be readable, writable, and executable. You’ll type this again and again
chmod 700 myscript.sh
Handy shortcuts
up(down)_key – scrolls through the command history
BASH Tricks: Auto-Completion
<something-incomplete> TAB – completes program_path/file_name
Taking control over the cursor (the pointer on the command line):
Ctrl+a # cursor to beginning of command line Ctrl+e # cursor to end of command line Ctrl-w # Cut last word Ctrl+k # cut to the end of the line Ctrl+y # paste content that was cut earlier (by Ctrl-w or Ctrl-k) history # shows all commands you have used recently
When specifying file names:
. (dot) – refers to the present working directory
Finding files, directories and applications
find -name "*pattern*" # searches for *pattern* in and below current directory find /usr/local -name "*blast*" # finds file names *blast* in specfied directory find /usr/local -iname "*blast*" # same as above, but case insensitive Additional useful arguments: -user <user name>, -group <group name>, -ctime <number of days ago changed> find ~ -type f -mtime -2 # finds all files you have modified in the last two days locate <pattern> # finds files and dirs that are written into update file which <application_name> # location of application whereis <application_name> # searches for executeables in set of directories grep pattern file # provides lines in 'file' where pattern 'appears', # if pattern is shell function use single-quotes: '>'grep -H pattern # -H prints out file name in front of pattern grep 'pattern' file | wc # pipes lines with pattern into word count wc
List directories and files
ls -al # shows something like this for each file/dir: drwxrwxrwx
# d: directory
# rwx: read write execute
# first triplet: user permissions (u)
# second triplet: group permissions (g)
# third triplet: world permissions (o)
More useful commands
df # disk space free -g # memory info in Megabytes uname -a # shows tech info about machine bc # command-line calculator (to exit type 'quit') wget <ftp> # file download from web /sbin/ifconfig # give IP and other network info ln -s original_file new_file # creates symbolic link to file or directory du -sh # displays disk space usage of current directory du -sh * # displays disk space usage of individual files/directories du -s * | sort -nr # shows disk space used by different directories/files sorted by size Process Management
Viewing text files
more <my_file> # views text, use space bar to browse, hit 'q' to exit less <my_file> # a more versatile text viewer than 'more', 'q' exits, 'G' end of text, 'g' beginning, '/' find forward, '?' find backwards cat <my_file> # concatenates files and prints content to standard output
Running and managing processes
top # view top consumers of memory and CPU (press 1 to see per-CPU statistics) who # Shows who is logged into system w # Shows which users are logged into system and what they are doing ps # Shows processes running by user ps -e # Shows all processes on system; try also '-a' and '-x' arguments ps aux | grep <user_name> # Shows all processes of one user ps ax --tree # Shows the child-parent hierarchy of all processes ps -o %t -p <pid> # Shows how long a particular process was running. # (E.g. 6-04:30:50 means 6 days 4 hours ...) Ctrl z <enter> # Suspend (put to sleep) a process fg # Resume (wake up) a suspended process and brings it into foreground bg # Resume (wake up) a suspended process but keeps it running in the background. Ctrl c # Kills the process that is currently running in the foreground kill <process-ID> # Kills a specific process kill -9 <process-ID> # NOTICE: "kill -9" is a very violent approach. It does not give the process any time to perform cleanup procedures. kill -l # List all of the signals that can be sent to a proccess kill -s SIGSTOP <process-ID> # Suspend (put to sleep) a specific process kill -s SIGCONT <process-ID> # Resume (wake up) a specific process renice -n <priority_value> # Changes the priority value, which range from 1-19,the higher the value the lower the priority, default is 10.
Special Charactes
Flavors of linux have a variety of special characters. When these are needed to be explicitly referred to, often one uses a \ to escape such as \” does not open an interpreted quote pair.
# Indicates what follows is to be ignored by the interpreter, except when the first line is #! as in #!/usr/bin/bash
echo "hello" # Don't echo this #hello echo "hello and # we do echo here" #hello and # we do echo here
Remember, | pipes the output into another program
history | tail # 504 echo "hello and # we do echo here" # 505 history | head # 506 history | head -3
> redirects standard out to a file
history > myfile.txt
; Ends the line without a ‘return’
echo hello; echo there
;; Terminates case
case "$variable" in abc) echo "\$variable = abc" ;; xyz) echo "\$variable = xyz" ;; esac
* Match all or wildcard
ls *.sh
" Preserves (from interpretation) most of the special characters from “String” as STRING.
echo "STRING $HOME" STRING /Users/dcraig
' Preserves all of the special characters as ‘”STRING”‘ as “STRING”
echo 'STRING $HOME' STRING $HOME
$ Indicates to interpolate variable
var1=5 var2=test echo $var1 # 5 echo $var2 # test
${variable} Same as $parameter, i.e., value of the variable parameter. In certain contexts, only the less ambiguous ${parameter} form works.
echo ${var1}
Control-V, Control Key, e.g. Press Control-v then press ‘retu4rn’ encodes the character for return
grep "text^M" # This must be tried to understand it
\ Literally interpret the following character such as \’
echo \' #Should print '
/ Separator for folder in the path
cd mypath/subdir
Archiving
Creating
tar -cvf my_file.tar mydir/ # Builds tar archive of files or directories. For directories, execute command in parent directory. Don't use absolute path. tar -czvf my_file.tgz mydir/ # Builds tar archive with compression of files or directories. For dirs, execute command in parent directory. zip -r mydir.zip mydir/ # Command to archive a directory (here mydir) with zip. tar -jcvf mydir.tar.bz2 mydir/ # Creates *.tar.bz2 archive
Viewing
tar -tvf my_file.tar tar -tzvf my_file.tgz
Extracting
tar -xvf my_file.tar tar -xzvf my_file.tgz gunzip my_file.tar.gz # or unzip my_file.zip, uncompress my_file.Z,or bunzip2 for file.tar.bz2 find -name '*.zip' | xargs -n 1 unzip # this command usually works for unzipping many files that were compressed under Windows tar -jxvf mydir.tar.bz2 # Extracts *.tar.bz2 archive
Part II: VIM
Exercise: Learning how to edit text with vim
VIM Editor Commands
Vim is an editor to create or edit a text file. There are two modes in vim. One is the command mode and another is the insert mode. In the command mode, the user can move around the file, delete text, etc, whereas in the insert mode, the user can insert text.
From command mode to insert mode type a/A/i/I/o/O ( see details below)
From insert mode to command mode type Esc (escape key)
Text Entry Commands (Used to start text entry)
- a Append text following the current cursor position
- A Append text to the end of the current line
- i Insert text before the current cursor position
- I Insert text at the beginning of the cursor line
- o Open up a new line following the current line and add text there
- O Open up a new line in front of the current line and add text there
The following commands are used only in the commands mode.
^F(CTRl F) Forward screenful^BBackward screenful^fOne page forward^bOne page backward^UUp half screenful^DDown half screenful$Move the cursor to the end of the current line0(zero) Move the cursor to the beginning of the current linewForward one wordbBackward one word
Exit Commands
:wqWrite file to disk and quit the editor:q!Quit (no warning):qQuit (a warning is printed if a modified file has not been saved)ZZSave workspace and quit the editor (same as :wq)
Text Deletion Commands
xDelete characterdwDelete word from cursor ondbDelete word backwardddDelete lined$Delete to end of lined^(d caret, not CTRL d) Delete to beginning of line
Yank (has most of the options of delete)– VI’s copy command
yyyank current liney$yank to end of the current line from cursorywyank from the cursor to end of the current word5yyank, for example, 5 lines
Paste (used after delete or yank to recover lines.)
ppaste below cursorPpaste above cursoruUndo last changeURestore lineJJoin next line down to the end of the current line
File Manipulation Commands
:wWrite workspace to original file:wfile Write workspace to named file.Repeat last commandrReplace one character at the cursor positionRBegin overstrike or replace mode � use ESC key to exit:g/pat1/s//pat2/greplace every occurrence of pattern1 (pat1) with pat2
Examples
Opening a New File
- Step 1 type vim filename (create a file named filename)
- Step 2 type i ( switch to insert mode)
- Step 3 enter text (enter your Ada program)
- Step 4 hit Esc key (switch back to command mode)
- Step 5 type :wq (write file and exit vim)
Part III: Scripting
BASH Scripting Core Concepts
Variables
A variable is a name given to for storing for a piece of information. There are two actions we may perform for variables: Setting a value for a variable; Reading the value for a variable. Variables may have their value set in a few different ways. The most common are to set the value directly and for its value to be set as the result of processing by a command or program.
When setting a variable we leave out the $ sign.
I=5
When referring to or reading a variable we place a $ sign before the variable name.
echo $I #returns 5
Special Variables
There are a few other variables that the system sets for you to use as well.
- $0 – The name of the Bash script.
- $1 – $9 – The first 9 arguments to the Bash script. (As mentioned above.)
- $# – How many arguments were passed to the Bash script.
- $@ – All the arguments supplied to the Bash script.
- $? – The exit status of the most recently run process.
- $$ – The process ID of the current script.
- $USER – The username of the user running the script.
- $HOSTNAME – The hostname of the machine the script is running on.
- $SECONDS – The number of seconds since the script was started.
- $RANDOM – Returns a different random number each time is it referred to.
- $LINENO – Returns the current line number in the Bash script.
- ${#var}Return the length of the variable var.
Example BASH #1: pointless.sh
pointless.sh
#!/bin/bash echo $VARIABLE VARIABLE=5 echo $HOSTNAME # prints name of server you are on. echo $VARIABLE #prints 5 VARIABLE=6 echo $VARIABLE #prints 6
Quotes, Backslashes
Quotes are important, and so are backslashes. Single quotes will treat every character literally. Double quotes will allow you to do substitution (that is include variables within the setting of the value). Backslashes literally interpret a special character such as $, so that \$MONEY is printed with the dollar sign “$MONEY”
Example BASH #2: quotequotes.sh
quotequotes.sh
#!/bin/bash echo $VARIABLE VARIABLE=5 echo $HOSTNAME # prints name of server you are on. echo $VARIABLE #prints 5 VARIABLE=6 echo $VARIABLE #prints 6
Export
Typically, variables are only available within the script, unless you export them to the environment. Then they are available to all scripts spawned from the script.
Example BASH #3: quotequotesexport.sh
quotequotesexport.sh
#!/bin/bash VARIABLE=4 export $VARIABLE pointless.sh # this script will start with $VARIABLE set to 4
Command Substitution In Bash
Command substitution allows us to take the output of a command or program (what would normally be printed to the screen) and save it as the value of a variable. To do this we place it within brackets, preceded by a $ sign – $(ls) would list directories
Example BASH script #3: processes.sh
process.sh
#!/bin/bash myvar=$( ps -ef | grep $USER) echo $myvar
Reading Standard Input
A program sends either to the standard output or standard err. We know that we can redirect these using the “>” sign. Also, a program takes in from standard input, and we know we can pipe results of one program as standard input into another program. Standard input could be single-purpose commands together to create a larger solution tailored to our exact needs. The ability to do this is one of the real strengths of Linux. It turns out that we can easily accommodate this mechanism with our scripts also. By doing so we can create scripts that act as filters to modify data in specific ways for us.
Bash accommodates piping and redirection by way of special files. Each process gets its own set of files (one for STDIN, STDOUT, and STDERR respectively) and they are linked when piping or redirection is invoked. Each process gets the following files:
STDIN – /dev/stdin
STDOUT – /dev/stdout
STDERR – /dev/stderr
Example BASH script #4: makeupper.sh
makeupper.sh # note use of tr program.
#!/bin/bash cat /dev/stdin | tr a-z A-Z # What does tr do? Look it up.
Math
let is a builtin function of Bash that allows us to do simple arithmetic. It follows the basic format:
+, -, /*, / addition, subtraction, multiply, divide var++ Increase the variable var by 1 var-- Decrease the variable var by 1 % Modulus (Return the remainder after division)
expr is similar to let except instead of saving the result to a variable it instead prints the answer. Unlike let you don’t need to enclose the expression in quotes. You also must have spaces between the items of the expression. It is also common to use expr within command substitution to save the output to a variable.
Example #5: math.sh
math.sh
#!/bin/bash let a=5+4 echo $a # 9 a=$( expr 10 - 3 ) echo $a # 7
Conditionals
If statements (and, closely related, case statements) are conditional code that operates only when an argument is true. Basically tests are within square brackets ( [ ] ) in the if statement, and can be TRUE or FALSE. We list common ones below. When we want to perform a certain set of actions if a statement is true, and another set of actions if it is false, we use the else. If we may have a series of conditions that may lead to different paths, we use if then else if.
If follows the format below:
if [ <some test> ]
then
<commands>
fi
or
if [ <some test> ]
then
<commands>
else
<other commands>
fi
or
if [ <some test> ]
then
<commands>
elif [ <some test> ]
then
<different commands>
else
<other commands>
fi
Anything between then and fi (if backward) will be executed only if the test (between the square brackets) is true.
PLEASE INDENT INSIDE THE IF BY 4 SPACES
Example #6: bigornot.sh
bigornot.sh
#!/bin/bash # Basic if statement if [ $1 -gt 100 ] then echo Hey that\'s a large number. pwd fi date
mb_or_kb.sh
#!/bin/bash
FILE=$1
if [ -f $FILE ]; then
SIZE=$(du -b "$FILE" | cut -f 1);
if [ $SIZE -ge 1000000 ]; then
MB=$(expr 1000000 \/ 100000);
echo "${MB}MB";
else
KB=$(expr 1000000 \/ 1000);
echo "${KB}KB";
fi
else
echo "File $FILE does not exist."
fi
Operators more!
Operators are important, and they are essentially mini functions. Some core operators for if statements
| Operator | Description |
|---|---|
| ! EXPRESSION | The EXPRESSION is false. |
| -n STRING | The length of STRING is greater than zero. |
| -z STRING | The lengh of STRING is zero (ie it is empty). |
| STRING1 = STRING2 | STRING1 is equal to STRING2 |
| STRING1 != STRING2 | STRING1 is not equal to STRING2 |
| INTEGER1 -eq INTEGER2 | INTEGER1 is numerically equal to INTEGER2 |
| INTEGER1 -gt INTEGER2 | INTEGER1 is numerically greater than INTEGER2 |
| INTEGER1 -lt INTEGER2 | INTEGER1 is numerically less than INTEGER2 |
| -d FILE | FILE exists and is a directory. |
| -e FILE | FILE exists. |
| -r FILE | FILE exists and the read permission is granted. |
| -s FILE | FILE exists and it’s size is greater than zero (ie. it is not empty). |
| -w FILE | FILE exists and the write permission is granted. |
| -x FILE | FILE exists and the execute permission is granted. |
Boolean
One can use boolean operators to join them together.
and: &&
or: ||
Example #7: useful.sh
useful.sh
#!/bin/bash
# and example
if [ -r $1 ] && [ -s $1 ]
then
echo "This file, $1 is useful."
fi
Loops
There are two loops, we will work on: for and while
While loops
While an expression is true, keep executing these lines of code. They have the following format:
while [ <some test> ]
do
<commands>
done
Example #8: count.sh
count.sh
#!/bin/bash
# Basic while loop
counter=1
while [ $counter -le 5 ]
do
echo $counter
((counter++))
done
echo All done
For Loop
For each of the items in a given list, perform the given set of commands. It has the following syntax.
for var in <list> do <commands> done
Example #8: forcount.sh
forcount.sh
#!/bin/bash
# Basic range in for loop
for value in {1..5}
do
echo $value
done
Example #9: forgenes.sh
fornames.sh
#!/bin/bash # Basic for loop
genes='PTEN KRAS TP53'
for gene in $genes do
echo $gene done
There are some lesser-used loops, such as select, do, and unless. Also, one can break and continue loops. They will not be reviewed in this course.
Functions
Functions in Bash mirror scripts – they provide mini-programs in someways. However, they are a core concept in most languages.
Functions can have the word function in front of them, but often we don’t as it is not required. It gets confusing, so it’s wise to indent. The form is generally:
function_name () {
VAR1=$1
VAR2=$2
echo $VAR1
echo $VAR2
}
Later we can shortcut these commands by typing
function_name("hello","goodbye") #prints hello goodbye
Passing Variables. In most programming languages, we frequently pass arguments to functions and they are assigned to variables listed in brackets. This is not the case in BASH and we have to manually assign them. They are provided to the functional internal scope as $1, $2, $3… . We may send data to the function in a similar way to passing command-line arguments to a script. We supply the arguments directly after the function name.
Returning. Most languages allow us to return a result. BASH doesn’t do this directly but does allow us to provide a status of whether there was a success or not using return.
Example script #10: whatgene.sh
whatgene.sh
#!/bin/bash
# Setting a return status for a function
print_gene () {
echo mygene $1
return 1
}
print_gene PTEN
print_gene Jupiter
Variable scope and reach
Scope refers to where a variable can be interpreted. BASH is again a little unique, and by default, variables are global within the script. They can be made local to a function. By default a variable is global. . To do that we use the keyword local in front of the variable the first time we set its value.
local var_name=<var_value>
Setting things local is good practice. This way variables are safer from being inadvertently modified by another part of the script which happens to have a variable with the same name (or vice versa).
Example script #11:
#!/bin/bash
# Showing scope
var_change () {
local var1='Initialized' #var1 is local to var_change function
echo Inside function: var1 is $var1 : var2 is $var2
var1='LOCAL:KRAS'
var2='LOCAL:PTEN'
}
var1='GLOBAL:KRAS'
var2='GLOBAL:PTEN'
echo Before function call: var1 is $var1 : var2 is $var2
var_change
echo After function call: var1 is $var1 : var2 is $var2
Servers
To log-in into your remote server, you typically use ssh from the command-line window. ITerm2 is the terminal program I will use. From the command line prompt:
ssh trgn510@trgn.usc.edu
You should be asked for your default password, and then given a prompt. You may get a warning if its the first time you have ever logged in. Your prompt should change slightly and indicate the server you have logged into. If you are unsure, type hostname that is a command that prints the name of the server you are on.
The first thing we should do is to change your password to something other than the default you were given. We do this using the command passwdType passwd with the prompt, and change it.
Copying files to and from servers
There are many ways to get files over to a server. Some are no longer used, and each server has its own rules that are set by the sysadmin. If you are using a MacOS, then it too can be a server, and you can open up the ability to open it up to the world. Some examples are telnet, scp, rsync, sftp, ftp and so forth. There are graphical tools which can aid in this process, but for now, we will ignore them. We will focus on rsync. We focus on rsync because scp only provides a cp like a method to copy files from one machine to a remote machine over a secure SSH connection – and better, rsync,provides syncronization options and pick up where things left off. rsync allows you to synchronize remote folders.
To copy data over to a computer, you typically are on a different computer. In your current MacOS allows you to create a file for transferring using the shell (I use Iterm2). Simply lets copy over our history. Open up a terminal, and type history | rev > ~/myreversehistory.txt. For fun, we have reversed the text using a pipe and then redirected into a file on the main home directory. Lets actually put it into a directory mkdir -p ~/myfolder and then mv that file into it: `mv ~/myreversehistory.txt ~/myfolder`
Now if we want to transfer that over we can type:
rsync –azvh myfolder davidwcr@trgn.usc.edu:~/
There are some options, I’m using. To better understand them you can look at the rsync manual, by typing man rsync though many web pages provide easier to understand options.
Exercise: AN EXAMPLE BASH SCRIPT
Example: Creating a BASH script
Retrieve some sample data, and unzip it
This data is GTF file or the formal gene and transcript reference file mapping all known genes and transcripts to build 37 of the human genome as defined by the Ensemble effort led out of the European Bioinformatics Institute. In this file, each line is a ‘feature’. You can see that an ‘exon’ is a feature, as a ‘gene’, ‘microRNA’, etc. This isn’t that helpful for our purpose which we define here as pulling out all o of the gene types and their positions in the Build 37.1. You can see that there would be quite a bit of data wrangling to pull out just the gene-names, and we will save this exercise for later in this assignment.
Typically, there is a reference file. You can learn more about the content here, and this is relevant for our efforts. http://ftp.ensembl.org/pub/release-75/gtf/homo_sapiens/README
wget http://ftp.ensembl.org/pub/release-75/gtf/homo_sapiens/Homo_sapiens.GRCh37.75.gtf.gz
Let’s unzip it
gunzip Homo_sapiens.GRCh37.75.gtf.gz
Let’s preview it
less is a program that allows us to browse a text file. Many people actually alias more=less in their .bashrc file. less is nice in that allows and recognizes many vim commands such as ‘/search’. To make our app, we will want a comma delimited file that has ‘feature’,’chr’,’start’,’stop’. There clearly is much more info here.
less Homo_sapiens.GRCh37.75.gtf
#!genome-build GRCh37.p13 #!genome-version GRCh37 #!genome-date 2009-02 #!genome-build-accession NCBI:GCA_000001405.14 #!genebuild-last-updated 2013-09 1 pseudogene gene 11869 14412 . + . gene_id "ENSG00000223972"; gene_name "DDX11L1"; gene_source "ensembl_havana"; gene_biotype "pseudogene"; 1 processed_transcript transcript 11869 14409 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; gene_name "DDX11L1"; gene_source "ensembl_havana"; gene_biotype "pseudogene"; transcript_name "DDX11L1-002"; transcript_source "havana"; 1 processed_transcript exon 11869 12227 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "1"; gene_name "DDX11L1"; gene_source "ensembl_havana"; gene_biotype "pseudogene"; transcript_name "DDX11L1-002"; transcript_source "havana"; exon_id "ENSE00002234944"; 1 processed_transcript exon 12613 12721 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "2"; gene_name "DDX11L1"; gene_source "ensembl_havana"; gene_biotype "pseudogene"; transcript_name "DDX11L1-002"; transcript_source "havana"; exon_id "ENSE00003582793"; 1 processed_transcript exon 13221 14409 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "3"; gene_name "DDX11L1"; gene_source "ensembl_havana"; gene_biotype "pseudogene"; transcript_name "DDX11L1-002"; transcript_source "havana"; exon_id "ENSE00002312635"; 1 transcribed_unprocessed_pseudogene transcript 11872 14412 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000515242"; gene_name "DDX11L1"; gene_source "ensembl_havana"; gene_biotype "pseudogene"; transcript_name "DDX11L1-201"; transcript_source "ensembl"; 1 transcribed_unprocessed_pseudogene exon 11872 12227 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000515242"; exon_number "1"; gene_name "DDX11L1"; gene_source
Let’s pull out a few fields
There are many ways to pull out a column and put into a file. One program that can do this is ‘cut’, another is ‘awk’. Here we use cut just for the purposes of showing different approaches. We use the -f3 which means to cut out the 3rd column presuming space deliminators. To see all of the options of cut, feel free to man cut.
cut -f3 Homo_sapiens.GRCh37.75.gtf | head
#!genome-build GRCh37.p13 #!genome-version GRCh37 #!genome-date 2009-02 #!genome-build-accession NCBI:GCA_000001405.14 #!genebuild-last-updated 2013-09 gene transcript exon exon exon
Let’s use awk instead for formatting
As we said, there are many ways to cut fields. awk , gawk, or nawk (different versions of awk) is one of the most common linux/unix/bash tools for wrangling data. In the example below, we print the 2nd, 1st, 4th, and 5th columns for those lines where 3rd column matches gene. By default, it splits by white space, but you can change this if you need to.
awk '$3 ~ /gene/ { print $2,$1,$4,$5}' Homo_sapiens.GRCh37.75.gtf | head
The $n refers to the nth field. The /text/ is how we match the lines we want to alter. In this case, if there is a “gene” in the 3rd column, we print some the $2, $1, $4, and $5 field for the file Homo_sapiens.GRCh37.75.gtf
pseudogene 1 11869 14412 pseudogene 1 14363 29806 lincRNA 1 29554 31109 lincRNA 1 34554 36081 pseudogene 1 52473 54936 pseudogene 1 62948 63887 protein_coding 1 69091 70008 lincRNA 1 89295 133566 lincRNA 1 89551 91105 pseudogene 1 131025 134836
Let’s make it comma delimited.
We could have added commas in awk. However, in this case, we show piping the results into sed. sed is essentially a search and replace tool replacing the space with ,. Remember that we are piping into ‘head to just get a preview. sed is based on regular expressions, another term for complex matching and substitution. sed generally works as s/find/replace/g. It becomes quite amazing how complex these can be. Please go to http://regex101.com to learn about how to make complex matches.
awk '$3 ~ /gene/ { print $2,$1,$4,$5}' Homo_sapiens.GRCh37.75.gtf | sed 's/ /,/g' | head
pseudogene,1,11869,14412 pseudogene,1,14363,29806 lincRNA,1,29554,31109 lincRNA,1,34554,36081 pseudogene,1,52473,54936 pseudogene,1,62948,63887 protein_coding,1,69091,70008 lincRNA,1,89295,133566 lincRNA,1,89551,91105 pseudogene,1,131025,134836
Redirecting into a file
We actually haven’t written any files yet. We are essentially running programs and then piping to head which prints only the first 5 lines or so. This provides us with a preview of what the multiple commands together are doing. This time lets actually put into a file by using the redirect or > into the file gene_dist.csv
awk '$3 ~ /gene/ { print $2,$1,$4,$5}' Homo_sapiens.GRCh37.75.gtf | sed 's/ /,/g' > gene_dist.csv
Give it a first line header line
For later steps, we really need a first line that tells us what our columns are. This is important for many reasons, particularly for R that we will use in a future step. We have decided to use sed for that as it also allows us to insert lines. Now, to be clear – I didn’t remember that at first, but upon googling, I found out how to do that functionality of sed. That is an important aspect that one does not need to remember every feature of every command.
sed -e "1ifeature,chr,start,end" gene_dist.csv > gene_dist_head.csv
feature,chr,start,end pseudogene,1,11869,14412 pseudogene,1,14363,29806 lincRNA,1,29554,31109 lincRNA,1,34554,36081 pseudogene,1,52473,54936 pseudogene,1,62948,63887 protein_coding,1,69091,70008 lincRNA,1,89295,133566 lincRNA,1,89551,91105
To get started we will go back to our directory.
cd ~/projects/mygraph ls -l
The results show:
total 832232 -rw-rw-r-- 1 tgrn510 tgrn510 2137228 Sep 7 05:56 gene_dist.csv -rw-rw-r-- 1 tgrn510 tgrn510 2137250 Sep 7 05:56 gene_dist_head.csv -rw-rw-r-- 1 tgrn510 tgrn510 847928511 Feb 7 2014 Homo_sapiens.GRCh37.75.gtf
We see our previous files, their permissions, and their size. Lets put what we just did in a script with a proper directory and versioning.
