David W Craig
David W. Craig, Ph.D.


David W Craig, Ph.D.
Professor and Co-Director
USC Institute for Translational Genomics
Overview
Dr. Craig is the vice chair of the Department of Translational Genomics at USC and the head of the Molecular Genomics Core at the Norris Comprehensive Cancer Center. Before coming to USC, he was the Deputy Director of Bioinformatics at TGen and Director of the Neurogenomics Division. With over 200 publications, my expertise is in translational genomics and bioinformatics.
Development of integrative genomic tools. In the past 15 years, Dr. Craig’s lab has used genomics to make experimental and computational tools that connect engineering, biotechnology, and clinical care. His group published one of the first methods for Illumina multiplexed sequencing in 2008 (Nature Methods). Integration of DNA and RNA is a primary focus, with papers and patents on cryptic splicing, fusion detection, X-skewing, and variant prioritization in cancer. The second area of focus in genomics has been untangling mixtures, which is very important in oncology. These approaches have led to many significant papers, including one of the most influential papers on data privacy problems (Homer et al., PLOS Genet, 2008).
Translational Genomics. Translating genomics from bench to bedside is the foundation of Dr. Craig’s research. He co-founded the CAP/CLIA-accredited Ashion Labs. Also, he helped start TGen’s Center for Rare Childhood Disorders (C4RCD.org) and got over a thousand families to join the study of diseases with unknown genetic causes. His group has worked to develop shared standards and datasets, serving as a co-PI on 1000 Genomes. (i) Neurological disorders. He has had collaborative publications in bipolar disorder (>45 pubs), Alzheimer’s disease (>15 pubs), and pediatric neurology (>30 pubs). As part of the Accelerated Medicine Partnership in Parkinson’s Disorder (AMP-PD), his group recently led the longitudinal analysis of over 8,500 transcriptomes from 1600 people. (ii) Oncology. His research in somatic heterogeneity and disease progression has led to collaborations developing genomic methods in oncology. His group was one of the first to study whole-genome and transcriptome profiling to treat metastatic triple-negative breast cancer.
Research
Biomedical Engineering – Applying to Human Health and Disease


Dr. David Craig’s Laboratory And Research Includes Integration of the Experimental Lab with the Dry Lab.
Bioinformatics. To improve outcomes in human disease, our lab focuses on the intersection of engineering, biotechnology, and clinical care. Genomic analysis is a vital part of precision medicine, and patient-by-patient analysis is now possible thanks to next-generation sequencing (NGS) technology. Patient data at the individual patient level has grown significantly during the past five years. To better understand how to choose treatments, we are currently at a unique point in time where we are redefining how we can look at individual-level data at the molecular, cellular, and systems levels. The following period comes before performing these measures over thousands of individual cells and builds on the emergence of single-molecule nucleic acid measurements from next-generation sequencing technology. This area of bioinformatics is the main topic of our study, which involves scientists, trainees, and students using sizable datasets both at the bench and in the dry lab.
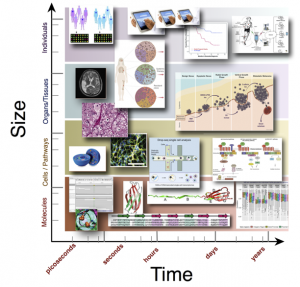

Analysis Requires Integration Across Dimensions
Moving at Multiple Scales
In the lab today, we are moving from the ability to generate genome-wide sequence data for a single patient to be able to generate single-molecule data, not just from one cell but from hundreds to thousands of cells together with cellular biology and different clinical measurements (such as from imaging and pathology). While the technology and data are forthcoming, considerable research and development are needed to optimally leverage and bridge together data spanning molecules, cells, and tissues. So, this academic program aims to research and develop ways to combine and model data from individual nucleic acid molecules and cells to system-level clinical measurements. This will help us learn more about diagnosing, treating, preventing, and stopping the spread of disease.
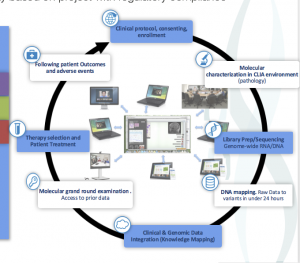

Iterative Integration of Data
Bioinformatics and Data Science
Bioinformatics, where the ideas of data science, clinical care, and biotechnology all meet, is one of our group’s most important research areas. We have several thrusts, primarily centered around the integration of multi-omic data.
Data Integration, Harmonization, and Establishment of Analysis Bioinformatics must build on processes for testing, validating, and integrating new tools as they come out and are needed by other projects and cores. Indeed, the analysis field for NGS continues to evolve and grow at a fantastic pace, producing new tools even while previous ones have just been put into production.
Data Integration, Harmonization & Establishment Of Analysis Approaches.Integration and modern software frameworks are critical for exploring and truly exploring big data. Graphs are moving beyond paper to dynamic and interactive activities that allow one to explore and interact with data. Our group uses full-stack development to support these, whereby terabytes of data are reduced and integrated into web portals. One such effort is through the Michael J. Fox Foundation, where we collaborate with others to develop portals to explore thousands of genomic DNA samples with RNA. This interactive framework allows for the inspection of different phenotypes and is built upon the D3.js, MongoDB, and BigTable frameworks that are easily deployable within controlled environments.
