David W. Craig, Ph.D.


David W Craig, Ph.D.
Professor and Co-Director
USC Institute for Translational Genomics
Overview
David W. Craig serves as Vice-Chair of USC’s new Department of Translational Genomics within the USC Keck School of Medicine. Dr. Craig’s expertise is in genomics, bioinformatics, and data analysis of high-throughput genomics data. His laboratory consists of both a wet-lab and dry-lab. Within his group, lab personnel has the opportunity to either specialize or become dual trained in bioinformatics and molecular biology.
His group pioneered cost-effective GWAS methods leading to genetic associations reported in Science, Nature Genetics, and New England Journal of Medicine. His publications include some of the most significant papers addressing the challenges of data sharing and data privacy. Since 2006, his team has been developing tools based on NGS beginning with publishing one of the first papers for targeted variant calling in humans (Craig et al Nature Methods, 2008). In the past 8 years, they have published and collaborated on over 60 NGS publications balanced between the wet and dry-labs. During this time, he has served in several international genomics projects, including as a PI on a U01 responsible for developing bioinformatic pipelines for the Phase I and Phase II portions of the 1000 Genomes Project.
With collaborators, his group was among the first to implement NGS in molecular profiling for cancer patient treatment recommendations in a feasibility study in metastatic triple-negative breast cancer (Craig et al., Mol Cancer Ther. 2013). Building upon this and other efforts his team developed an end-to-end platform for personalized medicine, NGS data management, analysis, and clinical genomic interpretation following CAP/CLIA guidelines. Within this framework, they completed analytical validation for integrated RNA/DNA analysis of tumor/normal sets. Community resources from these efforts included include a collaborative release of COLO829 tumor/normal sequencing reference sets. He also was a founding scientific director for TGen’s Center for Rare Childhood Disorders – a research clinic enrolling over 1000 individuals into a study developing integrative RNA/DNA approaches for identifying the germline genetic basis of disease.
Research
Biomedical Engineering - Applying to Human Health and Disease


Dr. David Craig's Laboratory And Research Includes Integration of the Experimental Lab with the Dry Lab.
Our lab focuses is focused on where engineering, biotechnology, and clinical care interface with a focus on impacting individual-level patient care. Genomics is playing a major role in precision medicine and next-generation sequencing (NGS) technologies have provided these capabilities on a patient level, and the last five years have seen burgeoning of individual-level patient data. Indeed, we have entered into a unique time fundamentally altering our ability to probe at a molecular, cellular, and systems-level scale the type individual-level data and our ability to assimilate this data for better understanding treatment decisions. The period that follows a revolution of single-molecule nucleic acid measurements from next-generation sequencing technologies and one that precedes making these measurements across thousands of cells individually. This is the focus of our research with trainees, students, and scientists working with large datasets both at the bench and the dry-lab.
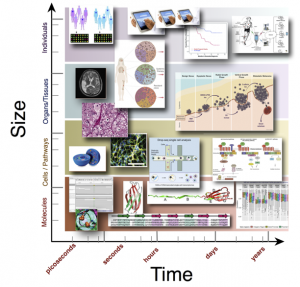

Analysis Requires Integration Across Dimensions
Moving at Multiple Scales
Today, we are moving from the ability to genome-wide sequence data for a single patient to in the coming year being able to generate in single-molecule data. For example, such as from exome sequencing, RNA-seq or methyl-Seq - but not just from one cell but hundreds to thousands of cells together with cellular biology and different clinical measurements!
While the technology and data are forthcoming, considerably research and development are needed to optimally leverage and bridge together data spanning molecules, cells, tissues. Consequentially, the goal of this academic program: to research and develop approaches for integrating and modeling data spanning from the individual nucleic acid molecules and cells to system-level clinical measurements in order to improve our ability to understand disease diagnosis, treatment, progression, and prevention.
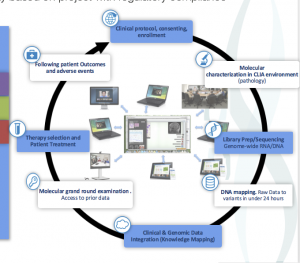

Iterative Integration of Data
BIOINFORMATICS AND DATA SCIENCE
One of the largest areas of research within our group is in the areas of bioinformatics, where principles of data science, clinical care, and biotechnology converge. We have several thrusts largely centered around the integration of multi-omic data.
Data Integration, Harmonization & Establishment Of Analysis Approaches. It is critical that the Bioinformatics Core has processes for testing, validating, and integrating new tools as they emerge and are needed by the other projects and cores. Indeed, the analysis field for NGS continues to evolve and grow at an amazing pace, producing new tools even while previous tools have just been placed in production. Moreover, the optimal approach for integrating and variant calling is typically specific to a study based on the technology, sample availability, and sample conditions. Change is inevitable and rapid, and we expect that there will be a need to implement new tools and analysis approaches even between the timing of submission and the time of commencing this study.
